ChromoTek RFP-Trap Agarose
RFP-Trap® Agarose is an affinity resin for IP of RFP-fusion proteins. It consists of an RFP Nanobody/ VHH coupled to agarose beads.
Specificity
mCherry, mPlum, mRFP
Applications
IP, CoIP, ChIP, RIP
Conjugate
Agarose
Type
Nanobody
256
Cat no : rta
Synonyms
Validation Data Gallery
Product Information
RFP-Trap® Agarose is an affinity resin for IP of RFP-fusion proteins. It consists of an RFP Nanobody/ VHH coupled to agarose beads.
| Description | Immunoprecipitation of RFP-fusion proteins and their interacting factors with anti-RFP Nanobody conjugated to beads.
• Reliable and robust pull-down of mCherry and other RFP-fusion proteins • Ready-to-use • No heavy & light antibody chains • Stable under harsh washing conditions • Suitable for downstream mass spec analysis • Works in samples from: mammals, plants, bacteria, yeast, insects etc. |
| Applications | IP, CoIP, ChIP, RIP |
| Specificity/Target | mRFP, mCherry, mRFPruby, mPlum, tagRFP, mKate2, mOrange, PA-mCherry, mScarlet For the complete list, please click here: Fluorescent protein specificity table |
| Binding capacity | 22.5 μg of recombinant RFP per 25 μL bead slurry |
| Conjugate | Agarose beads; bead size: ~ 90 µm (cross-linked 4 % agarose beads) |
| Elution buffer | SDS sample buffer 0.2 M glycine pH 2.5 |
| Wash buffer compatibility | 10 mM DTT, 4 M Urea, 2 M NaCl, 2 % Nonidet P40 Substitute, 1 % Triton X-100 |
| Type | Nanobody |
| Class | Recombinant |
| Host | Alpaca |
| Affinity (KD) | Dissociation constant KD of 5 nM |
| Compatibility with mass spectrometry | The RFP-Trap® is optimized for on-bead digestion. For the application note, please click here: On-bead digest protocol for mass spectrometry |
| RRID | AB_2631362 |
| Storage Buffer | 20% ethanol |
| Storage Condition | Shipped at ambient temperature. Upon receipt store at 4°C. Stable for one year. Do not freeze! |
Documentation
| SDS |
|---|
| SDS RFP-Trap Agarose (PDF) |
| Protocol |
|---|
| Protocol RFP-Trap Agarose (PDF) |
| RFP-Trap Specificity |
|---|
| Fluorescent protein specificity table (PDF) |
| Brochure |
|---|
| Product brochure (PDF) |
| Application notes |
|---|
| Chromatin Immunoprecipitation (ChIP) with RFP-tagged proteins |
| Split Fluorescent Protein Technology (PDF) |
| Trouble shooting |
|---|
| Troubleshooting guide immunoprecipitation (IP) (PDF) |
Publications
| Application | Title |
|---|---|
Science Reconstitution of autophagosome nucleation defines Atg9 vesicles as seeds for membrane formation. | |
Cell Exome capture reveals ZNF423 and CEP164 mutations, linking renal ciliopathies to DNA damage response signaling. | |
Cell Formation of NPR1 Condensates Promotes Cell Survival during the Plant Immune Response. | |
Cell A Defense Pathway Linking Plasma Membrane and Chloroplasts and Co-opted by Pathogens. | |
Reviews
The reviews below have been submitted by verified Proteintech customers who received an incentive forproviding their feedback.
FH Andrea (Verified Customer) (10-05-2023) | Works well for Co-IP with RFP tag.
|
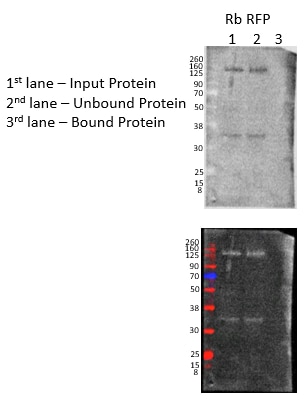
FH Julie (Verified Customer) (07-07-2021) | I was using this kit to try a co-IP. My IP protein is tagged to cherry, but the very first step of IP did not work so I could not test for an interaction with other protein candidates. The antibody I used works very well. As you can see, the protein of interest is in the input and unbound fractions - meaning the RFP-protein did not bind to the RFP-trap agarose beads. Disappointed. I used buffers that were in the range that was recommended. It would be helpful if exact starting amounts of ingredients for buffers were provided since I can't test multiple conditions with a sample that only 2 reactions. I'm sure there is a way to troubleshoot, but again, I can't test multiple conditions with just 2 reactions and brain tissue is precious.
 |

