ChromoTek GFP-Trap® Agarose
GFP-Trap® Agarose is an affinity resin for IP of GFP-fusion proteins. It consists of a GFP Nanobody/ VHH coupled to agarose beads.
Specificity
GFP and GFP derivates
Applications
IP, CoIP, ChIP, RIP
Type
Nanobody
Conjugate
Agarose
3793
Cat no : gta
Synonyms
Validation Data Gallery
Product Information
GFP-Trap® Agarose is an affinity resin for IP of GFP-fusion proteins. It consists of a GFP Nanobody/ VHH coupled to agarose beads.
| Description | Immunoprecipitation of GFP-fusion proteins and their interacting factors with anti-GFP Nanobody conjugated to beads.
• Fast, reliable & efficient one-step immunoprecipitation • Ready-to-use • No heavy & light antibody chains • Stable under harsh washing conditions • Suitable for downstream mass spec analysis • Works in samples from: mammals, plants, bacteria, yeast, insects etc. |
| Applications | IP, CoIP, ChIP, RIP |
| Specificity/Target | AcGFP, Clover, eGFP, Emerald, GFP, GFP5, GFP Envy, GFP S65T, mGFP, mPhluorin, PA-GFP, Superfolder GFP, TagGFP, TagGFP2, monomeric eGFP A206K, CFP, YFP, Citrine, eCitrine, eYFP, Venus, Ypet, BFP For the complete list, please click here: Fluorescent protein specificity table |
| Binding capacity | 25-30 µg of recombinant GFP per 25 µL bead slurry |
| Conjugate | Agarose beads; bead size: ~ 90 µm (cross-linked 4 % agarose beads) |
| Elution buffer | SDS sample buffer 0.2 M glycine pH 2.5 |
| Wash buffer compatibility | 1 mM DTT, 3 M Guanidinium•HCl, 8 M Urea, 2 M NaCl, 2 % Nonidet P40 Substitute, 1 % SDS, 1 % Triton X-100 |
| Type | Nanobody |
| Class | Recombinant |
| Host | Alpaca |
| Affinity (KD) | Dissociation constant KD of 1 pM |
| Compatibility with mass spectrometry | The GFP-Trap® is optimized for on-bead digestion. For the application note, please click here: On-bead digest protocol for mass spectrometry |
| RRID | AB_2631357 |
| Storage Buffer | 20% ethanol |
| Storage Condition | Shipped at ambient temperature. Upon receipt store at 4°C. Stable for one year. Do not freeze! |
Documentation
| SDS |
|---|
| SDS GFP-Trap Agarose |
| Protocol |
|---|
| Protocol GFP-Trap Agarose (PDF) |
| GFP-Trap 琼脂糖珠操作说明(中文) |
| GFP-Trap Specificity |
|---|
| Fluorescent protein specificity table |
| Trouble shooting |
|---|
| Troubleshooting guide immunoprecipitation (IP) |
| Brochure |
|---|
| GFP-Trap product brochure |
Publications
| Application | Title |
|---|---|
Cell Sensory circuitry controls cytosolic calcium-mediated phytochrome B phototransduction | |
Science Induction of lysosomal and mitochondrial biogenesis by AMPK phosphorylation of FNIP1 |
Reviews
The reviews below have been submitted by verified Proteintech customers who received an incentive forproviding their feedback.
FH Katie (Verified Customer) (01-12-2024) | These beads are wonderful! They gave us the cleanest IP I have ever seen. We are switching over to this product for all immunoprecipitations going forward.
|
FH Despoina (Verified Customer) (10-31-2023) | Our lab has been using the ChromoTek GFP-Trap® Agarose beads for a couple of years now in co-immunoprecipitation experiments, followed by western blotting. The product works well and produces clear blots.
|
FH Andrea (Verified Customer) (09-19-2023) | Works well well for co-immunoprecipitation.
|
FH Alessandro (Verified Customer) (01-19-2023) | great binding
|
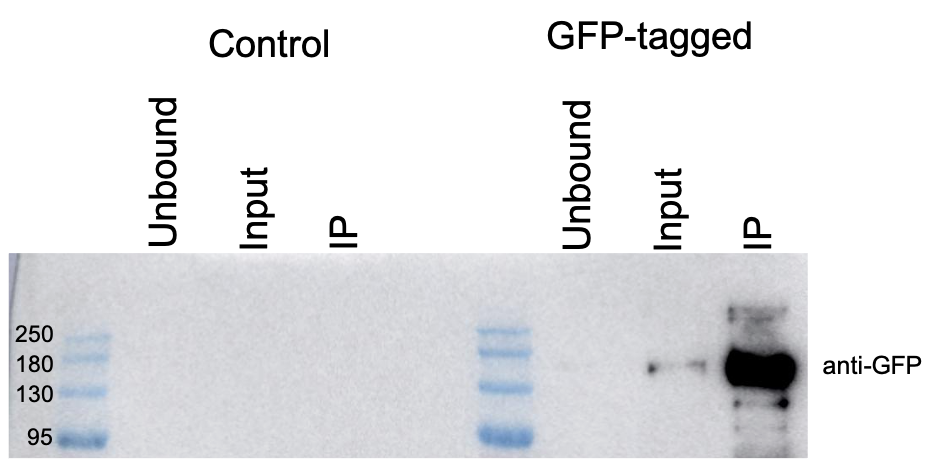
FH Liam (Verified Customer) (01-11-2023) | Easy to use and produces clean/reliable results. Over the past three years, the ChromoTek GFP-Trap® Agarose have become a core experimental resource for our lab.
 |
FH Rich (Verified Customer) (09-12-2022) | works well. A little on the pricey side but worth it.
|
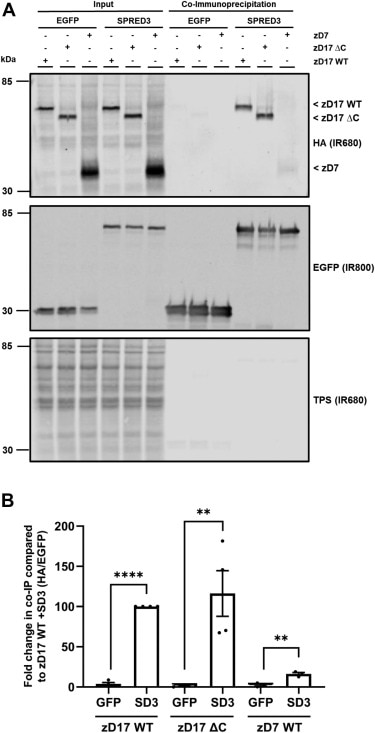
FH Aditi (Verified Customer) (08-17-2022) | The product works very well even with non-conventional model systems. We used it for co-immunoprecipitation and mass spectrometry analysis. We used custom buffers and the product worked efficiently. The whole protocol is also quicker than regular IP protocols. Highly recommend!
 |
FH Benedikt (Verified Customer) (08-17-2022) | We did a pull-down of a ~131 GFP-fusionprotein using the recommended buffers and incubation times (1h) and immediately pulled our protein-of-interest. Some non-bound protein was still visible, although this might be due to high amount of input lysate (500 µg).
|
FH Peter (Verified Customer) (07-04-2022) | Our lab has used the GFP-Trap Agarose beads on many occasions to successfully co-immunoprecipitate GFP fusion proteins for Mass Spectrometry and for RNA sequencing (RIP-Seq). We even managed to pulldown enough of one protein to detect it after the western blot on the PVDF membrane with amidoblack staining.
|
FH Cristina (Verified Customer) (06-10-2022) | we have been using this product for years and it's one of the fav in the lab
|
FH George (Verified Customer) (06-10-2022) | Used this for GFP immunoprecipitations from HeLa cells. Product worked as described and is much quicker and more effective than conventional immunoprecipitations. Would highly recommend.
|
FH Cristina (Verified Customer) (03-11-2022) | Great product. It is a daily must have in our lab.
|

