About Nanobodies
Nanobodies are powerful alpaca antibodies. ChromoTek engineers Nanobodies with carefully designed affinities, valencies, and specificities. Our innovative Nanobody derived products can be used in multiple research applications and as high potential building blocks for cancer immunoimaging, immunotherapy, and other therapeutic or diagnostic applications.

General and ChromoTek-specific advantages and benefits of alpaca Nanobodies for proteomics and cell biology. Three chapters are discussed:
- General advantages of alpaca antibodies
- Benefits for pulldown and purification
- Advantages for microscopy
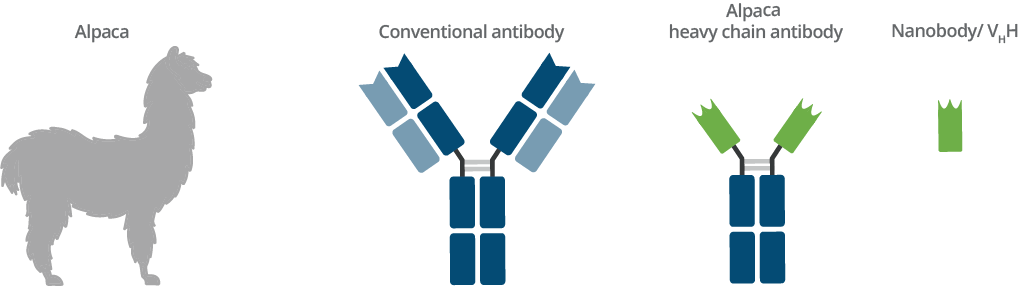
Origin of Nanobodies and comparison with conventional antibody
General advantages of alpaca Nanobodies
Camelids such as dromedaries, llamas, and alpacas possess an immune repertoire of three subclass IgG antibodies: IgG1, IgG2, and IgG3. IgG1 is a conventional IgG composed of two heavy chains and two light chains. IgG2 and IgG3 are heavy chain only IgGs antibodies (HCAbs) that can be distinguished by their hinge regions. These HCAbs lack the CH1 domain of the heavy chain and are devoid of any light chain. The antigen binding domain of a heavy chain only IgG is called VHH or Nanobody. VHHs have several general advantages; ChromoTek’s Nanobodies and Nanobody-based research tools, however, have even more advantages: their high level of performance, characterization, and validation.
Camelids comprise so-called old-world camelids, i.e. Camelus bactrianus and Camelus dromedarius and new-world camelids, i.e. Lama pacos, Lama glama, and Lama vicugna.
Small-size
- Our Nanobodies are 15 kDa in size and about 10-fold smaller than conventional IgG antibodies (150 kDa)
- Higher labelling density
- Higher image resolution because of very small 2 nm epitope label displacement
Epitope-access
- Good epitope access even in crowed cellular environments
- Considerable better penetration into tissues, organs, and animals
Robust binding molecules:
- Chemical and thermal stability
- Long shelf life time
- Room temperature shipment
Reliable & reproducible VHHs:
- Recombinant production
- Monoclonal
- Robust conjugation
- Quality controlled
Validated affinity reagents:
- Sequence and structure validated
- Function validated
- Published in more than 1,600 scientific publications
- Virtual no production batch to batch variation
Benefits for pulldown and purification
For immunoprecipitation, affinity purification, and additional biochemical applications, the ChromoTek Nanobodies and Nano-Traps (= Nanobody coupled to agarose bead) do provide multiple advantages for superior performance:
Single band purification
- No contamination of IgG antibody heavy and light chains
- Highly specific binding
- Low background
High stability
- High chemical stability allows for very harsh buffer and washing conditions, e.g. up to 8 M urea. ChromoTek’s Nanobodies and Nano-Traps are individually tested for binding in detergents, salt, reducing agents, and chaotropes (see specifications of individual Nano-Trap).
- High thermal stability allows binding at elevated temperatures of up to 83 °C (see specifications of individual Nano-Trap).
High affinity
- The kinetic properties of our Nanobodies have been thoroughly determined. Very high affinities with dissociation constants (KD) in a range from 1 pM to 7 nM have been determined. These high affinities enable the effective binding of proteins even at low concentration, i.e. at endogenous protein expression level (see specifications of individual Nano-Trap).
Ready to use
- ChromoTek’s VHHs are covalently bound to agarose and magnetic agarose beads
- Nano-Traps are ready for pulldown or affinity purification
- High binding capacity (see specification of individual Nano-Trap).
Low background
- Advanced coupling to matrix to reduce non-specific binding
Proven
- More than 2,200 scientific publications
Advantages for Microscopy
ChromoTek’s Nanobodies that are coupled to fluorescent dyes are called Nano-Boosters or Nano-Labels. These provide multiple advantages for superior imaging performance in multiple immunofluorescence applications such as epifluorescence, confocal, and super resolution microscopy:
Small-size
- Our Nanobodies are about 10-fold smaller than conventional IgG antibodies and 2 nm in size
- 100 µg of Nanobody/ Nano-Booster corresponds to 1,000 µg of conventional IgG antibody
Higher labelling density
- More molecules of Nano-Boosters/ Nano-Labels than by bulky IgG molecules can access the antigens in crowded cellular environments.
Higher image resolution
- Higher image resolution because of very small 2 nm epitope label displacement of a monovalent Nano-Booster/ Nano-Label, which is at least 10 times less than (a complex) of conventional antibodies/ secondaries
- Ideal for super resolution microscopy applications
Epitope-access
- Considerable better penetration into tissues, organs, and animals:
- ChromoTek Nano-Boosters GFP-Booster and RFP-Booster have been used to label whole genetic modified mice after clearing procedures
No clustering
- Monovalent Nanobodies do not cluster unlike conventional IgG antibodies, which are bivalent and thus may form large clusters
Avidity effects from bivalent formats
- Bivalent Nanobody formats do gain considerably from avidity effects and provide better labelling performance when needed
How does ChromoTek validate their alpaca single domain antibodies?
ChromoTek utilizes genetic strategies and comparison with independent antibodies for the validation of our VHHs against fluorescent proteins and peptide tags:
- In the genetic approach, VHHs are tested in their target application (immunoprecipitation, immunofluorescence and/or Western blot) both on cell lines that express and do not express their cognate fluorescent protein or peptide tag.
- In addition, our Nanobodies are benchmarked with established conventional antibodies.
Our Nanobodies are always sequenced; in several cases, we even know their crystal structure. Furthermore, we thoroughly characterize and validate our monoclonal Nanobodies. Their recombinant production in combination with high QC standards ensures reliable and stable alpaca single domain antibody products virtually without lot-to-lot variations.
Discussion of antibody validation
The validation and reliable production of antibodies are intensively discussed in life sciences both in academia and pharmaceutical industry (Bradbury and Plückthun 2015; Baker 2015). This debate complains about insufficient antibody quality and, consequently, wasted research time and funds and seeks options to resolve these issues by standardizing antibodies and their production.
A comprehensive set of guidelines for the validation of antibodies was recently published in “A proposal for validation of antibodies” by the International Working Group for Antibody Validation (M. Uhlen et al. 2016). The working groups suggests:
Five conceptual pillars for the validation of antibodies
- genetic strategies
- orthogonal approaches
- comparison with an independent antibody
- tagged protein expression
- IP-MS
Of these, at least one, better more approaches should be used to validate an antibody properly. ChromoTek utilizes in fact two approaches for validation: genetic strategies and comparison with independent antibodies for the validation of our VHHs against fluorescent proteins and peptide tags.
A short journey into the discovery of the Nanobody/ VHH technology
What’s so special about alpaca antibodies and how were they discovered? Alpacas aren’t typical lab animals, so these are legitimate questions...
As is often the case with important discoveries, chance helped scientists at the Free University of Brussels in the late 1980s. As Michael Gross remembers the story: During a practical course, a couple of biology students were to extract antibodies from human blood serum. They were not overly excited, on the one hand because they were concerned that the samples might be contaminated with HIV, on the other hand because this type of experiment had already been done numerous times before and the result was well documented in their text books. Their tutors then offered to sacrifice a few mice instead – not a very popular choice either. Eventually, a few liters of frozen dromedary serum were discovered in the lab freezer – this exotic example inspired the students to start working on the antibody separation.
In addition to the usual distribution of immunoglobins, they also discovered a group of smaller antibodies that did not correspond to anything known to science. This might have ended in obscurity, had not two researchers, Raymond Hamers and Cecile Casterman, investigated the matter more deeply. They did not believe that this species were just degraded variants of the “real” antibodies and therefore started to characterize them in more detail. Eventually, it became clear that they had discovered a new class of antibodies that were devoid of light chains and had a single antigen recognizing domain. These antibodies were later found in different camelid species, including llamas and alpacas. If you would like to learn more about the captivating story of this discovery, which includes travels to Morocco, a stolen camel and help from a Sheikh, you may read Michael Gross’ book “The birds, the Bees and the Platypuses”.
Based on their structure, these peculiar camelid antibodies have been named Heavy Chain Antibodies (hcAb), as they are composed of heavy chains only and are devoid of light chains. HcAbs are not found in other mammals except in pathological cases. In 1995, Greenberg and colleagues found similar hcAbs in nurse sharks (Greenberg et al., 1995), but evolutionary analysis showed that camelid and shark hcAbs evolved independently (Nguyen et al., 2002). There are many speculations about the evolutionary driving force for the emergence of heavy chain antibodies in such distantly related species. A plausible explanation could be that, unlike conventional (comparably large) antibodies, these small single domain antigen binding fragments allow the targeting of otherwise inaccessible epitopes, e.g. catalytic centers of enzymes (Flajnik et al., 2011).
In the absence of light chains, the fragment-antigen-binding (Fab) part of these antibodies is reduced to a single domain. Therefore, hcAbs belong to the class of single domain antibodies (sdAbs). The single domain is called VHH (variable heavy domain of heavy chain antibodies) domain or Nanobody. The VHH domain contains a complete antigen binding site and is the smallest functional antigen binding fragment (around 15 kDa - only one tenth the size of a conventional antibody).
In fact, the many advantages of this novel class of antibodies range from research applications to drug development. The first approval of a Nanobody-based drug was in 2018, when Caplacizumab developed by Ablynx, now part of Sanofi, was launched for the treatment of acquired thrombotic thrombocytopenic purpura (aTTP).
In general, Nanobodies can be readily selected and produced in bacteria, ensuring their virtually unlimited supply in consistent quality. In contrast to conventional antibodies, Nanobodies are also exceptionally stable, withstanding conditions of extreme temperatures, chaotropic reagents, detergents, glycerol, salt, reducing conditions, and pH. At ChromoTek, we thoroughly characterize our single domain antibody fragment derived products by function and structure: we prove applications and determine the chemical and thermal stability of every VHH individually. The stability of our products is outstanding: we have tested some Nanobody preparations that were more than five years old: they were still functional, with little or no loss of activity.